Difference between revisions of "Pioneer/Imaging Lab 1"
(→Example 11: Chips!) |
(→Example 12: Chip Edges!) |
||
| Line 289: | Line 289: | ||
=== Example 12: Chip Edges!=== | === Example 12: Chip Edges!=== | ||
<syntaxhighlight lang=python> | <syntaxhighlight lang=python> | ||
| − | + | import matplotlib.image as mpimg | |
imgo = mpimg.imread("amandajones_chips_unsplash.jpg") | imgo = mpimg.imread("amandajones_chips_unsplash.jpg") | ||
img = imgo[::10, ::10, :] | img = imgo[::10, ::10, :] | ||
| Line 308: | Line 308: | ||
fig, ax = plt.subplots(num=k+2, clear=True) | fig, ax = plt.subplots(num=k+2, clear=True) | ||
edgevals[:,:,k] = sig.convolve2d(img[:,:,k], h, "valid") | edgevals[:,:,k] = sig.convolve2d(img[:,:,k], h, "valid") | ||
| − | aplot1 = ax.imshow(edgevals[:,:,k], cmap=plt.cm.gray, vmin=- | + | aplot1 = ax.imshow(edgevals[:,:,k], cmap=plt.cm.gray, vmin=-1020, vmax=1020) |
ax.axis('equal') | ax.axis('equal') | ||
ax.set(title=names[k]+" Edges as Gray") | ax.set(title=names[k]+" Edges as Gray") | ||
| Line 316: | Line 316: | ||
fig, ax = plt.subplots(num=k+5, clear=True) | fig, ax = plt.subplots(num=k+5, clear=True) | ||
| − | aplot2 = ax.imshow(edgevals[:,:,k], cmap=plt.cm.gray, vmin=0, vmax= | + | aplot2 = ax.imshow(edgevals[:,:,k], cmap=plt.cm.gray, vmin=0, vmax=1020) |
ax.axis('equal') | ax.axis('equal') | ||
ax.set(title="Absolute " + names[k]+" Edges as Gray") | ax.set(title="Absolute " + names[k]+" Edges as Gray") | ||
Revision as of 00:48, 6 August 2021
This page serves as a supplement to the first Digital Image Processing Lab for the Summer 2021 Pioneer Program.
Contents
- 1 Corrections / Clarifications to the Handout
- 2 Running Python
- 3 Scikit Image
- 4 Links
- 5 Examples
- 5.1 Example 1: Black & White Images
- 5.2 Example 2: Simple Grayscale Images
- 5.3 Example 3: Less Simple Grayscale Images
- 5.4 Example 4: Building an Image
- 5.5 Example 5: Exploring Colors
- 5.6 Example 6: 1D Convolution
- 5.7 Example 7: 1D Convolution Using "same"
- 5.8 Example 8: 11x11 Blurring
- 5.9 Example 9: Make No Assumptions
- 5.10 Example 10: Basic Edge Detection and Display
- 5.11 Example 11: Chips!
- 5.12 Example 12: Chip Edges!
Corrections / Clarifications to the Handout
- None yet
Running Python
You need to have Anaconda installed, which includes the Spyder IDE and all the modules that we need for this assignment.
Scikit Image
See Pioneer21/Lecture03 for more details about using the scikit-image module.
Links
- 2D Convolution description: HTTP://www.songho.ca/dsp/convolution/convolution2d_example.HTML
- Kernel page on Wikipedia: https://en.wikipedia.org/wiki/Kernel_(image_processing)
- Test Card for Exercise 8: https://en.wikipedia.org/wiki/Test_card
Examples
The following sections will contain both the example programs given in the lab as well as the image or images they produce. You should still type these into your own version of Python to make sure you are getting the same answers. These are provided so you can compare what you get with what we think you should get.
Example 1: Black & White Images
a = np.array([
[1, 0, 1, 0, 0],
[1, 0, 1, 0, 1],
[1, 1, 1, 0, 0],
[1, 0, 1, 0, 1],
[1, 0, 1, 0, 1]])
fig, ax = plt.subplots(num=1, clear=True)
ax.imshow(a, cmap=plt.cm.gray)
ax.set_axis_off()
fig.tight_layout()
fig.savefig("PS21DI1E1Plot1.png")
fig, ax = plt.subplots(num=2, clear=True)
aplot2 = ax.imshow(a, cmap=plt.cm.gray)
fig.colorbar(aplot2)
fig.tight_layout()
fig.savefig("PS21DI1E1Plot2.png")
Example 2: Simple Grayscale Images
bx, by = np.meshgrid(range(255), range(255))
fig, ax = plt.subplots(num=1, clear=True)
aplot1 = ax.imshow(bx, cmap=plt.cm.gray)
fig.colorbar(aplot1)
fig.tight_layout()
fig.savefig("PS21DI1E2Plot1.png")
fig, ax = plt.subplots(num=2, clear=True)
aplot2 = ax.imshow(by, cmap=plt.cm.gray)
fig.colorbar(aplot2)
fig.tight_layout()
fig.savefig("PS21DI1E2Plot1.png")
Example 3: Less Simple Grayscale Images
x, y = np.meshgrid(np.linspace(0, 2*np.pi, 201),
np.linspace(0, 2*np.pi, 201));
z = np.cos(x)*np.cos(2*y);
fig, ax = plt.subplots(num=1, clear=True)
zplot = ax.imshow(z, cmap=plt.cm.gray)
ax.axis('equal')
fig.colorbar(zplot)
fig.tight_layout()
fig.savefig("PS21DI1E3Plot1.png")
Notice how the use of ax.axis('equal') made the image look like a square since it is 201x201 but also caused the display to be filled with whitespace as a result of the figure size versus the image size.
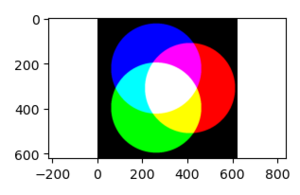
Example 4: Building an Image
rad = 100
delta = 10
x, y =np.meshgrid(range((-3*rad-delta),(3*rad+delta)+1),
range((-3*rad-delta),(3*rad+delta)+1))
rows, cols = x.shape
dist = lambda x, y, xc, yc: np.sqrt((x-xc)**2+(y-yc)**2)
venn_img = np.zeros((rows, cols, 3));
venn_img[:,:,0] = (dist(x, y, rad*np.cos(0), rad*np.sin(0)) < 2*rad);
venn_img[:,:,1] = (dist(x, y, rad*np.cos(2*np.pi/3), rad*np.sin(2*np.pi/3)) < 2*rad);
venn_img[:,:,2] = (dist(x, y, rad*np.cos(4*np.pi/3), rad*np.sin(4*np.pi/3)) < 2*rad);
fig, ax = plt.subplots(num=1, clear=True)
ax.imshow(venn_img)
ax.axis('equal')
fig.tight_layout()
fig.savefig("PS21DI1E4Plot1.png")
Example 5: Exploring Colors
nr = 256
nc = 256
x, y = np.meshgrid(np.linspace(0, 1, nc),
np.linspace(0, 1, nr))
other = 0.5;
palette = np.zeros((nr, nc, 3))
palette[:,:,0] = x
palette[:,:,1] = y
palette[:,:,2] = other
fig, ax = plt.subplots(num=1, clear=True)
ax.imshow(palette)
ax.axis('equal')
fig.tight_layout()
fig.savefig("PS21DI1E5Plot1.png")
Example 6: 1D Convolution
x = np.array([1, 2, 4, 8, 7, 5, -1])
h = np.array([1, -1])
y = sig.convolve(x, h)
print("x: {}\nh: {}\ny: {}".format(x, h, y))
x: [ 1 2 4 8 7 5 -1]
h: [ 1 -1]
y: [ 1 1 2 4 -1 -2 -6 1]
Example 7: 1D Convolution Using "same"
x = np.array([1, 2, 4, 8, 7, 5, -1])
h = np.array([1, -1])
y = sig.convolve(x, h, "same")
print("x: {}\nh: {}\ny: {}".format(x, h, y))
x: [ 1 2 4 8 7 5 -1]
h: [ 1 -1]
y: [ 1 1 2 4 -1 -2 -6]
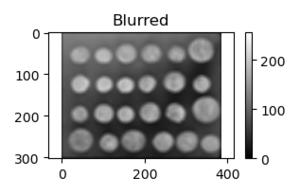
Example 8: 11x11 Blurring
x = ski.data.coins()
h = np.ones((11, 11))/11**2;
y = sig.convolve2d(x, h, 'same');
fig, ax = plt.subplots(num=1, clear=True)
aplot1 = ax.imshow(x, cmap=plt.cm.gray, vmin=0, vmax=255)
ax.axis('equal')
ax.set(title='Original')
fig.colorbar(aplot1)
fig.tight_layout()
fig.savefig("PS21DI1E8Plot1.png")
fig, ax = plt.subplots(num=2, clear=True)
aplot2 = ax.imshow(y, cmap=plt.cm.gray, vmin=0, vmax=255)
ax.axis('equal')
ax.set(title='Blurred')
fig.colorbar(aplot2)
fig.tight_layout()
fig.savefig("PS21DI1E8Plot2.png")
Example 9: Make No Assumptions
x = np.array([1, 2, 4, 8, 7, 5, -1])
h = np.array([1, -1])
y = sig.convolve(x, h, "valid")
print("x: {}\nh: {}\ny: {}".format(x, h, y))
x: [ 1 2 4 8 7 5 -1]
h: [ 1 -1]
y: [ 1 2 4 -1 -2 -6]
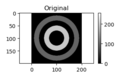
Example 10: Basic Edge Detection and Display
x, y = np.meshgrid(np.linspace(-1, 1, 200),
np.linspace(-1, 1, 200));
z1 = (.7<np.sqrt(x**2+y**2)) * (np.sqrt(x**2+y**2)<.9)
z2 = (.3<np.sqrt(x**2+y**2)) * (np.sqrt(x**2+y**2)<.5)
zimg = 100*z1+200*z2;
fig, ax = plt.subplots(num=1, clear=True)
aplot1 = ax.imshow(zimg, cmap=plt.cm.gray, vmin=0, vmax=255)
ax.axis('equal')
ax.set(title='Original')
fig.colorbar(aplot1)
fig.tight_layout()
fig.savefig("PS21DI1E9Plot1.png")
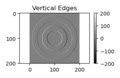
hx = np.array([[1, -1], [1, -1]])
edgex = sig.convolve2d(zimg, hx, 'valid')
fig, ax = plt.subplots(num=2, clear=True)
aplot2 = ax.imshow(edgex, cmap=plt.cm.gray)
ax.axis('equal')
ax.set(title='Vertical Edges')
fig.colorbar(aplot2)
fig.tight_layout()
fig.savefig("PS21DI1E9Plot2.png")
hy = np.array([[1, 1], [-1, -1]])
edgey = sig.convolve2d(zimg, hy, 'valid')
fig, ax = plt.subplots(num=3, clear=True)
aplot3 = ax.imshow(edgey, cmap=plt.cm.gray)
ax.axis('equal')
ax.set(title='Horizontal Edges')
fig.colorbar(aplot3)
fig.tight_layout()
fig.savefig("PS21DI1E9Plot3.png")
edges = np.sqrt(edgex**2 + edgey**2)
fig, ax = plt.subplots(num=4, clear=True)
aplot4 = ax.imshow(edges, cmap=plt.cm.gray)
ax.axis('equal')
ax.set(title='Edges')
fig.colorbar(aplot4)
fig.tight_layout()
fig.savefig("PS21DI1E9Plot4.png")
Example 11: Chips!
For this example, save the image at right to a file called "amandajones_chips_unsplash.jpg" in the same folder as Python is running. This image comes from, Amanda Jones' Unsplash page and is free to use for non-commercial use.
import matplotlib.image as mpimg
from matplotlib.colors import ListedColormap
img = mpimg.imread("amandajones_chips_unsplash.jpg")
fig, ax = plt.subplots(num=1, clear=True)
ax.imshow(img)
ax.axis('equal')
ax.set(title='Original')
fig.tight_layout()
fig.savefig("PS21DI1E11Plot1.png")
vals = np.linspace(0, 1, 256)
names = ['Red', 'Green', 'Blue']
for k in range(3):
fig, ax = plt.subplots(num=k+2, clear=True)
aplot1 = ax.imshow(img[:,:,k], cmap=plt.cm.gray, vmin=0, vmax=255)
ax.axis('equal')
ax.set(title=names[k]+" as Gray")
fig.colorbar(aplot1)
fig.tight_layout()
fig.savefig("PS21DI1E11Plot{}.png".format(k+2))
fig, ax = plt.subplots(num=k+5, clear=True)
mycmvals = np.zeros((256,4))
mycmvals[:,k] = vals
mycmvals[:,3] = 1
mycm = ListedColormap(mycmvals)
aplot2 = ax.imshow(img[:,:,k], cmap=mycm, vmin=0, vmax=255)
ax.axis('equal')
ax.set(title=names[k]+" as "+names[k])
fig.colorbar(aplot2)
fig.tight_layout()
fig.savefig("PS21DI1E11Plot{}.png".format(k+5))
Example 12: Chip Edges!
import matplotlib.image as mpimg
imgo = mpimg.imread("amandajones_chips_unsplash.jpg")
img = imgo[::10, ::10, :]
fig, ax = plt.subplots(num=1, clear=True)
ax.imshow(img)
ax.axis('equal')
ax.set(title='Scaled Down Original')
fig.tight_layout()
fig.savefig("PS21DI1E12Plot1.png")
vals = np.linspace(0, 1, 256)
names = ['Red', 'Green', 'Blue']
h = np.array([[1, 0, -1],[ 2, 0, -2], [ 1, 0, -1]])
edgevals = np.zeros((img.shape[0]-h.shape[0]+1,
img.shape[1]-h.shape[1]+1,
3))
for k in range(3):
fig, ax = plt.subplots(num=k+2, clear=True)
edgevals[:,:,k] = sig.convolve2d(img[:,:,k], h, "valid")
aplot1 = ax.imshow(edgevals[:,:,k], cmap=plt.cm.gray, vmin=-1020, vmax=1020)
ax.axis('equal')
ax.set(title=names[k]+" Edges as Gray")
fig.colorbar(aplot1)
fig.tight_layout()
fig.savefig("PS21DI1E12Plot{}.png".format(k+2))
fig, ax = plt.subplots(num=k+5, clear=True)
aplot2 = ax.imshow(edgevals[:,:,k], cmap=plt.cm.gray, vmin=0, vmax=1020)
ax.axis('equal')
ax.set(title="Absolute " + names[k]+" Edges as Gray")
fig.colorbar(aplot2)
fig.tight_layout()
fig.savefig("PS21DI1E12Plot{}.png".format(k+5))
fig, ax = plt.subplots(num=8, clear=True)
edgevals2 = (edgevals + abs(edgevals).max()) / 2 / abs(edgevals).max()
ax.imshow(edgevals2)
ax.axis('equal')
ax.set(title="Colorful Edges")
fig.tight_layout()
fig.savefig("PS21DI1E12Plot8.png")
fig, ax = plt.subplots(num=9, clear=True)
edgevals3 = np.linalg.norm(edgevals, ord=2, axis=2)
ax.imshow(edgevals3, cmap=plt.cm.gray)
ax.axis('equal')
ax.set(title="Absolute Edges")
fig.tight_layout()
fig.savefig("PS21DI1E12Plot9.png")